The data used for this project consist of Next Generation Sequencing (NGS) data from experiments I've been running on genetic switches. These switches are sequences of DNA which result in messenger RNA which can self-cleave, effectively reducing gene expression. A library of approximately 200,000 different, but closely related, switches were constructed and then used to construct DNA which was then transcribed in vitro into RNA. These molecules were then sequenced using NGS to give a dataset consisting of over 100 million reads of up to 200 nucleotides of DNA each. As they represent the inital library plus any mutations that accrue during the synthesis, measurement, and sequencing, these sequences are all very similar, often varying in only a few positions.
Here are some example sequences of the switches from the actual sequencing data (actually a shorter part of the sequencing results with only the switch itself retained):
GCTGTCACCGGAATACCAGCATCGTCTTGATGCCCTTGGCAGAGGACGAAACAGC GCTGTCACCGGAATACCAGCATCGTCTTGATGCCCTTGGCAGGGACGAAACAGC GCTGTCACCGGATGTGCTTTCCGGTCTGATGAGTCCATACCAGCATCGTCTTGATGCCCTTGGCAGGGACGAAACAGC ... GCTGTCACCGGATGTGCTTTCCGGTCTGATGAGTCCATACCAGCATCGTCTTGATGCCCTTGGCAGGGACGAAACAGC
Sonification of these data could be interesting in terms of being able to detect patterns in the sequences that might not be obvious from visual inspection.
These data were first taken and mapped directly to sound as follows:
This was implemented in ChucK using a data reader (SeqReader) and a player (playPulses) reading the data file seq and resulting in the following output: foo
Clearly, the output is quite "noisy", not only not sounding aesthetically pleasing, but not very revealing as to any patterns in the data.
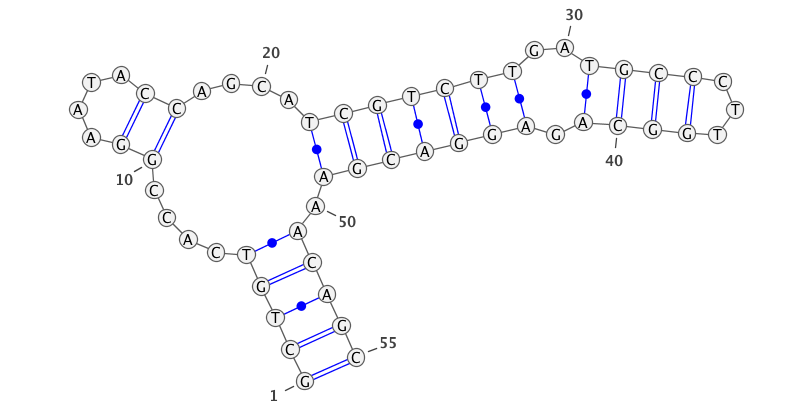
An interesting property of RNA (and of DNA) is that is folds up in a structured way creating a so-called secondary structure. This structure is primarily driven by the interaction of the bases to form pairs; the A bases form bonds with the T (actually Uracil, but the corresponding DNA base is Thymine), and the C bases pair with G. For example, the first sequence above forms a secondary structure that can be illustrated as:

The sections that are paired together, such as the GCTGT at positions 1-5 with the ACAGC at positions 51-55 are actually double helixes, whereas the intervening parts are single-stranded less structured RNA. The secondary structure is driven by the actual sequence, but there are multiple possible solutions for a given sequence. For example, the above sequence could also fold into the structure:

In fact, the same molecule of RNA may be rapidly switching between the various possibly secondary structure, albeit with different probabilities of each one depending on the energy required to maintain that conformation.
In the sequencing data I'm analyzing, this secondary structure of the RNA is one of the most important characteristics, determining how the molecule interacts with other molecules. Thus, mapping the structure rather than the actual sequence onto sound is attractive. To do this, I created an instrument whose pitch is governed by the structure. At a regular time interval, the invididual bases of a structure are traversed and these rules are followed:
Thus, each time a hairpin (a paired helix capped by a loop) is encountered, the pitch steps up according to the length of the helix, is held for a number of beats equal to the loop size, with amplitude decreasing for each note, and the stepped back down as the helix is exitted. By the time the entire structure is read, the pitch has returned to the base note of the scale.
The code for doing this consists of two parts -- a reader similar to the sequence reader, but returning an entire structure (one line of the data file) each time it is called. The file data is simply a string of 3 symbols, "(", ".", and ")". The left parenthesis represents a base of a pairing that is encountered the first time, the right parenthesis is the matching second base, and dots are for unpaired bases. Thus the coding for the top structure above would be:
(((((....((....))....(((((((..((((...)))).))))))).)))))
The reader is FoldReader which implements an open() and a next() method. The sound generator is playFold and the data files used are folds-1 (1 fold per sequence) and folds-10 (10 stochastic foldings per sequence). Here are some outputs based on different settings within playFold: